Description
Abbreviation: FNSGp(PGM)
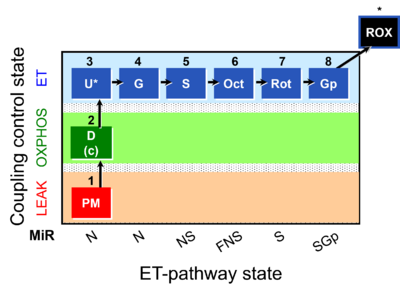
Reference: A: SUIT 1 is the RP1 - MiPNet21.06 SUIT RP
SUIT number: 1
MitoPedia concepts:
SUIT protocol,
SUIT A
- MitoPedia: SUIT - SUIT reference protocol RP1
- SUIT-category: FNSGp(PGM)
- SUIT protocol pattern: orthogonal
Harmonized SUIT protocols
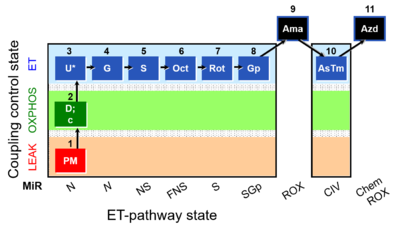
- SUIT RP1:1PM;2D:2c;3U;4G;5S;6Oct;7Rot;8Gp;9Ama;10AsTm;11Azd
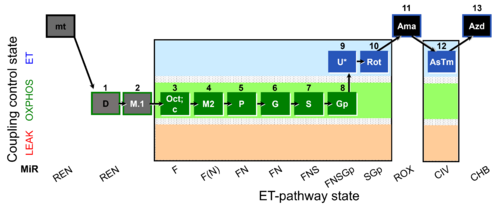
- SUIT RP2:1D;2M.1;3Oct;3c;4M2;5P;6G;7S;8Gp;9U;10Rot;11Ama;12AsTm;13Azd
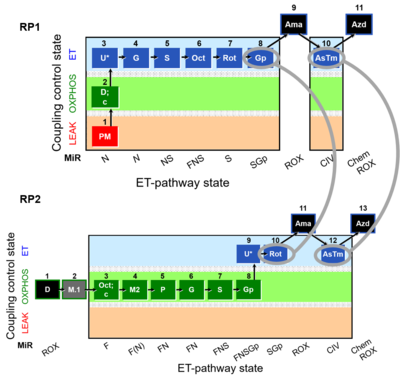 Harmonization between RP1 and RP2
Harmonization between RP1 and RP2
RP1 can be assessed in cells after the selective plasma membrane permeabilization. Digitonin is a mild detergent that permeabilizes plasma membranes. The optimum effective digitonin concentration for complete plasma membrane permeabilization of cultured cells can be determined directly in a respirometric protocol (see Digitonin). The use of intact cells allows the possibility to evaluate respiration in two different modules: intact cells and permeabilized cells. ROUTINE respiration (ce1) is obtained before digitonin is employed, whereas after the use of digitonin we can address differentet Electron transfer-pathway states.
- SUIT RP1 in cells:ce1;1Dig;1PM;2D:2c;3U;4G;5S;6Oct;7Rot;8Gp;9Ama;10Tm;11Azd
- SUIT RP2 in cells:ce1;1Dig;1D;2M.1;3Oct;3c;4M2;5P;6G;7S;8Gp;9U;10Rot;11Ama;12AsTm;13Azdd
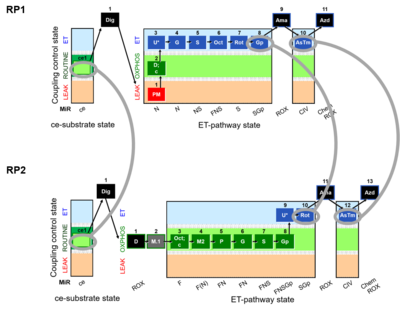 Harmonization between RP1 and RP2 in cells
Harmonization between RP1 and RP2 in cells
In PBMCs and PLTs the reference protocol RP1 is performed in the absense of Oct: SUIT 1 for PBMCs and PTLs
- SUIT RP1 for PBMCs and PLTs:1PM;2D;2c;3U;4G;5S;6Rot;7Gp;8Ama;9AsTm;10Azd
- SUIT RP2:1D;2M.1;3Oct;3c;4M2;5P;6G;7S;8Gp;9U;10Rot;11Ama;12AsTm;13Azd
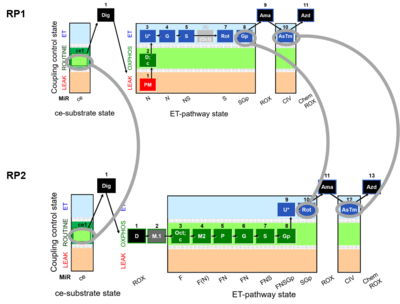 Harmonization between RP1 and RP2 in PBMC and PTL cells
Harmonization between RP1 and RP2 in PBMC and PTL cells
SUIT O2 DLP for SUIT 1 (RP1) (DatLab 7.3)
Updating this page (2018-06-13) for DatLab 7.3 DLP files