Wasilewski 2016 Abstract IOC115
| MitoRUSH, microscopy approach to mitochondrial protein import in mammalian cells. |
Link: Mitochondr Physiol Network 21.10
Wasilewski M, Mohanraj K, Chacinska A (2016)
Event: IOC115
The majority of mitochondrial proteins is encoded by nuclear DNA and synthesized in the cytosol, thus import machineries are required to allow proper localization of proteins into multiple subcompartments of mitochondria. There are five major import pathways, which specialize in the import of different classes of proteins. Proteins of the mitochondrial matrix as well as many proteins of the inner mitochondrial membrane, which are characterized by an N terminal signal known as a presequence, are imported on the presequence translocase of the inner membrane (TIM23) pathway. Most of our knowledge about TIM23 pathway stems from yeast were the main components and mechanisms governing TIM23 were characterized. Despite a high level of conservation of import machineries between yeast and Metazoa our understanding of TIM23 pathway in mammals is still rudimental. For example mammalian genomes encode for additional isoforms of some key subunits of TIM23 complex like TIM17A and B, which are both expressed ubiquitously but form separate sub-populations of the TIM23.
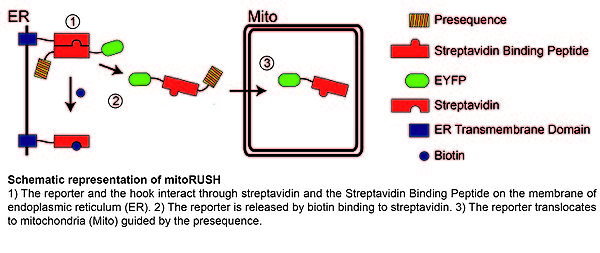
The progress in our understanding of mitochondrial protein import in mammalian cells is hindered by the lack of proper tools, which could substitute for in organello import strategy successful in yeast studies where large scale mitochondria isolation is feasible. In contrast studying mammalian cells often allows only limited amount of material. We propose a novel, microscopy-based approach named mitoRUSH (retention using selective hooks), which allows observation of mitochondrial import in living mammalian cells. MitoRUSH is based on expression of two fusion proteins called a “hook” and a “reporter”. The hook contains a transmembrane domain targeted to the ER membrane and a streptavidin domain exposed to the cytoplasm. The reporter consists of streptavidin binding peptide, a presequence and a fluorescent tag. Expressed together these proteins interact through streptavidin and a streptavidin binding peptide, which leads to accumulation of the reporter on the ER membrane. The reporter can be synchronously released from the hook by biotin applied to the cells and its translocation to mitochondria can be followed in a confocal microscope. We propose that this method can be applied to study mitochondrial protein import along the TIM23 pathway in vivo.
Labels:
Affiliation
Intern Inst Mol Cell Biol,Warsaw, Poland. - [email protected]
Figure 1
Figure 1.