Template:Oroboros News: Difference between revisions
From Bioblast
Beno Marija (talk | contribs) No edit summary |
No edit summary |
||
| (45 intermediate revisions by 4 users not shown) | |||
| Line 2: | Line 2: | ||
<div style="font-size:100%;font-weight:bold;padding:0.2em;padding-right: 0.4em;padding-left: 0.4em;background-color:#eeeeee;border-bottom:1px solid #aaaaaa;text-align:left;"> | <div style="font-size:100%;font-weight:bold;padding:0.2em;padding-right: 0.4em;padding-left: 0.4em;background-color:#eeeeee;border-bottom:1px solid #aaaaaa;text-align:left;"> | ||
[[Image:OROBOROS-solo.png|80px|right|link=Oroboros symbol |Oroboros symbol]] | [[Image:OROBOROS-solo.png|80px|right|link=Oroboros symbol |Oroboros symbol]] | ||
''' | '''Oroboros News''' | ||
</div> | </div> | ||
<div style="background-color:#ffffff;padding-top:0.2em;padding-right: 0.4em;padding-bottom: 0.2em;padding-left: 0.4em;"> | <div style="background-color:#ffffff;padding-top:0.2em;padding-right: 0.4em;padding-bottom: 0.2em;padding-left: 0.4em;"> | ||
''' | [[Image:MitoPedia.jpg|60px|link=http://www.bioblast.at/index.php/MitoPedia|MitoPedia]] '''''New in'' MitoPedia: [[International Union of Pure and Applied Chemistry, IUPAC |100 years of IUPAC]]''' - towards a '''common language'''. | ||
[[ | [[Image:O2k-brief.png|60px|link=http://wiki.oroboros.at/index.php/O2k-brief|O2k-brief]] '''''High-Resolution Respirometry. Respiration media''''' - [http://wiki.oroboros.at/images/7/7f/Wollenman_2017_Plos_One_O2k-brief.pdf Wollenman 2017]''' | ||
<br/> | |||
<br/> | |||
[[Image:Logo OROBOROS-DatLab.jpg|left|120px||link=http://wiki.oroboros.at/index.php/DatLab |DatLab]] | |||
<big><big>'''DatLab 7.3 release'''</big></big> | |||
We are proud to release DatLab 7.3, optimized for the Oroboros O2k-Series H, and enhancing applications of all O2k configurations. The software includes numerous new features: | |||
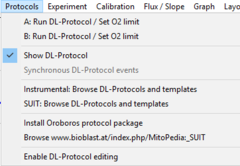
[[File:Protocol-menu.png|right|240px|link=Run DL-Protocol/Set O2 limit]] | |||
* '''Calibration and flux analysis''' – simplified for O2k-FluoRespirometer channels and add-on O2k-Modules. | |||
* '''DatLab Protocols:''' Both (1) instrumental protocols for quality control and (2) experimental substrate-uncoupler-inhibitor (SUIT) protocols guide the user step-by-step through the experiment, with programmed '''Events''' (titrations) and '''Marks''' (sections of the experiment for statistical analysis). | |||
* '''Browse DL-Protocols and templates:''' In a library of standardized protocols, each SUIT protocol is explained by an application example and supported by a template for data analysis. | |||
* '''Lower O2 limit:''' Respiration becomes oxygen dependent below a critical oxygen concentration. An automatic warning alerts the user to avoid hypoxia. | |||
* '''Graphs:''' Standard graph layouts can be selected, adjusted, and saved as User-defined layouts, to view O2k-MultiSensor experiments and export graphs for publication. Titration-induced peaks and spikes can be deleted. | |||
* '''Mark statistics:''' New statistical features are available for numerical display of results from DL-Protocol Marks and user-defined marks: median, average, SD, outlier index. These data can be exported for further analysis. | |||
* ''More details:'' [[MiPNet21.16_DatLab_7_Innovations |DatLab 7.3 Innovations]]''' | |||
<br/> | <br/> | ||
</div> | </div> | ||
Latest revision as of 15:22, 9 May 2019
 New in MitoPedia: 100 years of IUPAC - towards a common language.
New in MitoPedia: 100 years of IUPAC - towards a common language.
 High-Resolution Respirometry. Respiration media - Wollenman 2017
High-Resolution Respirometry. Respiration media - Wollenman 2017
DatLab 7.3 release
We are proud to release DatLab 7.3, optimized for the Oroboros O2k-Series H, and enhancing applications of all O2k configurations. The software includes numerous new features:
- Calibration and flux analysis – simplified for O2k-FluoRespirometer channels and add-on O2k-Modules.
- DatLab Protocols: Both (1) instrumental protocols for quality control and (2) experimental substrate-uncoupler-inhibitor (SUIT) protocols guide the user step-by-step through the experiment, with programmed Events (titrations) and Marks (sections of the experiment for statistical analysis).
- Browse DL-Protocols and templates: In a library of standardized protocols, each SUIT protocol is explained by an application example and supported by a template for data analysis.
- Lower O2 limit: Respiration becomes oxygen dependent below a critical oxygen concentration. An automatic warning alerts the user to avoid hypoxia.
- Graphs: Standard graph layouts can be selected, adjusted, and saved as User-defined layouts, to view O2k-MultiSensor experiments and export graphs for publication. Titration-induced peaks and spikes can be deleted.
- Mark statistics: New statistical features are available for numerical display of results from DL-Protocol Marks and user-defined marks: median, average, SD, outlier index. These data can be exported for further analysis.
- More details: DatLab 7.3 Innovations