Difference between revisions of "Coupling/pathway control diagram"
| Line 2: | Line 2: | ||
|abbr=CPCD | |abbr=CPCD | ||
|description=[[File:SUIT-nomenclature.jpg|300px|right|SUIT protocols]] | |description=[[File:SUIT-nomenclature.jpg|300px|right|SUIT protocols]] | ||
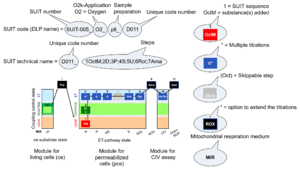
'''Coupling-pathway control diagrams''' illustrate the step-by-step substrate-uncoupler-inhibitor titrations in a [[SUIT protocol]]. The | '''Coupling-pathway control diagrams''' illustrate the respiratory '''states''' obtained step-by-step in substrate-uncoupler-inhibitor titrations in a [[SUIT protocol]]. Each step (the next state) is defined by an initial state and a [[metabolic control variable]], ''X''. The respiratory states are shown by boxes. ''X'' is usually the titrated substance in a SUIT protocol. If ''X'' (such as ADP or an uncoupler) exerts '''coupling control''', then a transition is induced between two [[coupling control state]]s. If ''X'' (such as pyruvate, succinate, or rotenone) exerts '''pathway control''', then a transition is induced between two [[pathway control state]]s. The type of metabolic control (''X'') is shown by arrows linking two respiratory states, with vertical arrows indicating coupling control, and horizontal arrows indicating pathway control. [[Marks in DatLab]] define the section of an experimental trace in a given [[respiratory state]] in the SUIT protocol. [[Events in DatLab]] define the titration of ''X'' in the SUIT protocol. The specific sequence of coupling control and pathway control steps defines the [[SUIT protocol pattern]]. The coupling-pathway control diagrams define the [[categories of SUIT protocols]] (see Figure). | ||
|info=[[Gnaiger 2014 MitoPathways]] | |info=[[Gnaiger 2014 MitoPathways]] | ||
}} | }} | ||
| Line 8: | Line 8: | ||
|mitopedia concept=MiP concept, SUIT concept | |mitopedia concept=MiP concept, SUIT concept | ||
}} | }} | ||
Communicated by [[Gnaiger Erich]] 2016-06-07, edited 2016-10-31. | |||
== See also == | == See also == | ||
Revision as of 10:33, 31 October 2016
- high-resolution terminology - matching measurements at high-resolution
Coupling/pathway control diagram
Description
Coupling-pathway control diagrams illustrate the respiratory states obtained step-by-step in substrate-uncoupler-inhibitor titrations in a SUIT protocol. Each step (the next state) is defined by an initial state and a metabolic control variable, X. The respiratory states are shown by boxes. X is usually the titrated substance in a SUIT protocol. If X (such as ADP or an uncoupler) exerts coupling control, then a transition is induced between two coupling control states. If X (such as pyruvate, succinate, or rotenone) exerts pathway control, then a transition is induced between two pathway control states. The type of metabolic control (X) is shown by arrows linking two respiratory states, with vertical arrows indicating coupling control, and horizontal arrows indicating pathway control. Marks in DatLab define the section of an experimental trace in a given respiratory state in the SUIT protocol. Events in DatLab define the titration of X in the SUIT protocol. The specific sequence of coupling control and pathway control steps defines the SUIT protocol pattern. The coupling-pathway control diagrams define the categories of SUIT protocols (see Figure).
Abbreviation: CPCD
Reference: Gnaiger 2014 MitoPathways
MitoPedia concepts:
MiP concept,
SUIT concept
Communicated by Gnaiger Erich 2016-06-07, edited 2016-10-31.